Rodent MRI Bias Field Correction
rodbfc.py is a Python module for performing bias field correction on rodent MRI images. This tool utilizes a pre-trained model to correct intensity inhomogeneities caused by the bias field present in MRI scans of rodents.
Features
Corrects bias field artifacts in rodent MRI images.
Easy-to-use command-line interface.
Uses a pre-trained model for accurate correction.
Installation
To use rodbfc.py, you’ll need to have Python 3 installed. Also
please install monai, pytorch, and nilearn python libraries.
You can install the module by cloning this repository. Pretrained models
are available in models directory, named according to date and time when
they were trained.
Usage
rodbfc.py -i input_filename -m model_filename -o output_filename [-b bias_filename] [--device DEVICE]
Arguments:
-h, --help: Show the help message and exit.-ior--input: Input filename (Uncorrected MRI image filename).-mor--model: Model file (Trained model .pth file).-o
or–output`: Output filename (Bias corrected MRI image filename).-b
or–bias`: Bias field filename (optional).--device: Device to use for computation (default: “cuda” if available, else “cpu”).
Example:
rodbfc.py -i input_image.nii.gz -m model_weights.pth -o output_image_corrected.nii.gz
Example output:
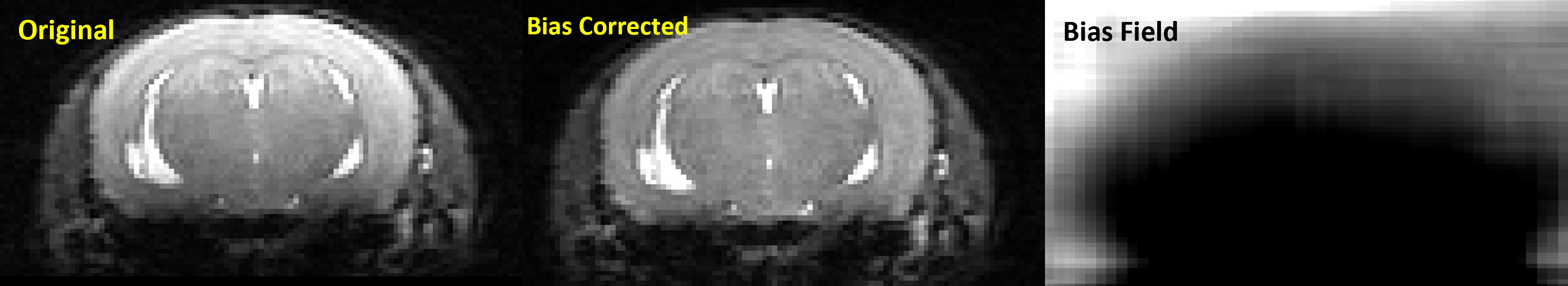
rodbfc
Training the model
A pretrained model is included in models directory. If you want to train
a new model, you can use the main_training.ipynb notebook.
License
- This project is licensed under the GNU General Public License v2.0 only
(GPL-2.0-only) License - see the LICENSE file for details.
Contributing
Contributions are welcome! Please contact ajoshi@usc.edu for further discussion.
Issues and Support
If you encounter any issues or need assistance, please create a Jira Issue in our repository.
Acknowledgments
This tool was developed by Anand A Joshi and Ronald Salloum. This project is supported by NIH Grant R01-NS121761 (PIs: David Shattuck and Allan MacKenzie-Graham).