1. MRI Processing Tools
1.1. Objective
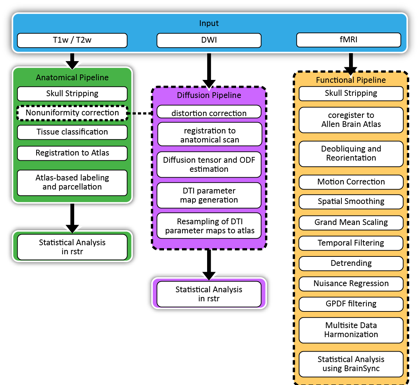
We are developing a set of image processing tools for rodent MRI to enable the assembly of analysis workflows. These tools include tissue classification, structural segmentation, registration to an atlas volume, labeling of neuroanatomical structures, and generation of structural models (cortex and subcortical areas). We will develop tools for processing diffusion MRI to generate parameter maps, tensor models and orientation distribution functions, and to perform diffusion tractography. We will also develop tools for processing fMRI data. These tools will provide the basis for rapidly constructing MRI analysis workflows for anatomical, diffusion, and functional MRI in rodent models.
1.2. Anatomical MRI
1.2.1. Brain Extraction
Identification of the brain in preclinical MRI is an important processing step in many imaging workflows. We have developed three tools for this purpose.
1.2.1.1. Mouse Brain Extractor (MBE)
Mouse Brain Extractor is a deep-learning-based method for segmenting the brain from mouse T2- and T1-weighted MRI data. Our method builds upon the SwinUNETR model (Hatamizadeh, 2021) and incorporates global positional encodings (GPEs). The addition of GPEs help boost segmentation accuracy in processing images at their native resolutions.
For more details, please see our preprint (Kim et al., 2024). Code and documentation are available from the Mouse Brain Extractor repository.
1.2.1.2. MouseBSE
MouseBSE is an adaptation of BrainSuite’s Brain Surface Extractor (BSE), which we developed previously for brain extraction in human MRI. BSE has been useful for non-human data, and the mouseBSE program captures some of the specifications that we frequently employ to make BSE work on rodent imaging.
1.2.1.3. maskbackgroundnoise
maskbackgroundnoise is a tool that we developed for ex-vivo imaging where the non-brain tissue has been physically removed prior to imaging. While these images do not have a skull to strip, differentiating brain tissue from background noise to identify the brain region of interest may still be challenging. This tools uses a combination of automatic thresholding and mathematical morphology to remove background voxels from the image.
1.2.2. Bias Field Correction
rodbfc.py is a Python module for performing bias field correction on rodent MRI images. This tool utilizes a pre-trained model to correct intensity inhomogeneities caused by the bias field present in MRI scans of rodents.
Corrects bias field artifacts in rodent MRI images.
Easy-to-use command-line interface.
Uses a pre-trained model for accurate correction.
1.2.3. Tissue Classification
1.2.4. Image Registration and Labeling
1.2.5. README
This README would normally document whatever steps are necessary to get your application up and running.
Quick summary
Version
Summary of set up
Configuration
Dependencies
Database configuration
How to run tests
Deployment instructions
Writing tests
Code review
Other guidelines
Repo owner or admin
Other community or team contact
1.3. Diffusion MRI
1.4. Functional MRI
1.4.1. README
This README would normally document whatever steps are necessary to get your application up and running.
Quick summary
Version
Summary of set up
Configuration
Dependencies
Database configuration
How to run tests
Deployment instructions
Writing tests
Code review
Other guidelines
Repo owner or admin
Other community or team contact
1.5. Atlases
We have assembled a set of anatomical mouse atlases for use in performing group studies and region-of-interest (ROI) analysis.
1.6. Comprehensive Workflow